蛋白质组的全局 pIs
细胞全局蛋白质组 pI 图的变化取决于氨基酸的总电荷,并对蛋白质的结构和特性具有重要意义。
普遍认为原核基因组具有两个最大的双峰形状,一个在酸性pH值下主要对应于溶解的蛋白质(细胞质蛋白或分泌蛋白),另一种在膜蛋白的碱性pH值下,具有细胞内碱性(带正电荷)结构域以促进质子动力的产生。在这两个峰之间,有一个最小的中性值,对应于细胞内的pH值(如下图)。
蛋白质氨基酸组成和 pI 水平的显着变化提供了一种工具来预测培养物或宏基因组组装基因组(MAG)的首选栖息地。
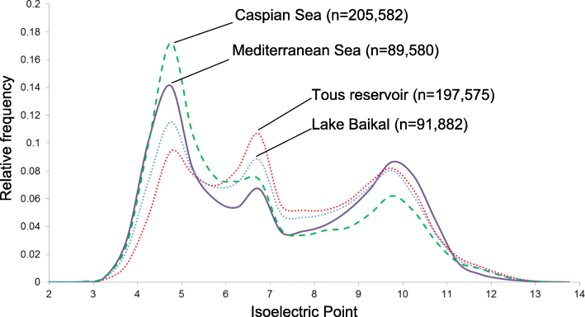
Pedro J. et al., 2019, Microbiome
安装EMBOSS
- 下载
1 | wget ftp://emboss.open-bio.org/pub/EMBOSS/emboss-latest.tar.gz |
- 安装
1 | # 解压 |
输入文件
输入文件为含有一条或多条氨基酸序列的FASTA格式文件。
计算氨基酸序列的各特征数据
逐个文件计算
1 | pepstats -sequence F01_bin.1.faa -outfile F01_bin.1.out |
:::primary 参数解析
Standard (Mandatory) qualifiers:
- [-sequence] seqall Protein sequence(s) filename and optional
format, or reference (input USA) - [-outfile] outfile [*.pepstats] Pepstats program output file
- [-sequence] seqall Protein sequence(s) filename and optional
Advanced (Unprompted) qualifiers:
- -aadata datafile [Eamino.dat] Amino acid properties
- -mwdata datafile [Emolwt.dat] Molecular weight data for amino
acids - -pkdata datafile [Epk.dat] Values of pKa for amino acids
- -[no]termini boolean [Y] Include charge at N and C terminus
- -mono boolean [N] Use monoisotopic weights
:::
批量计算与pI提取并输出为相对丰度
1 | #!/usr/bin/perl |
选择性忽略 (这是我自己用的)
1 | #!/usr/bin/perl |
可视化
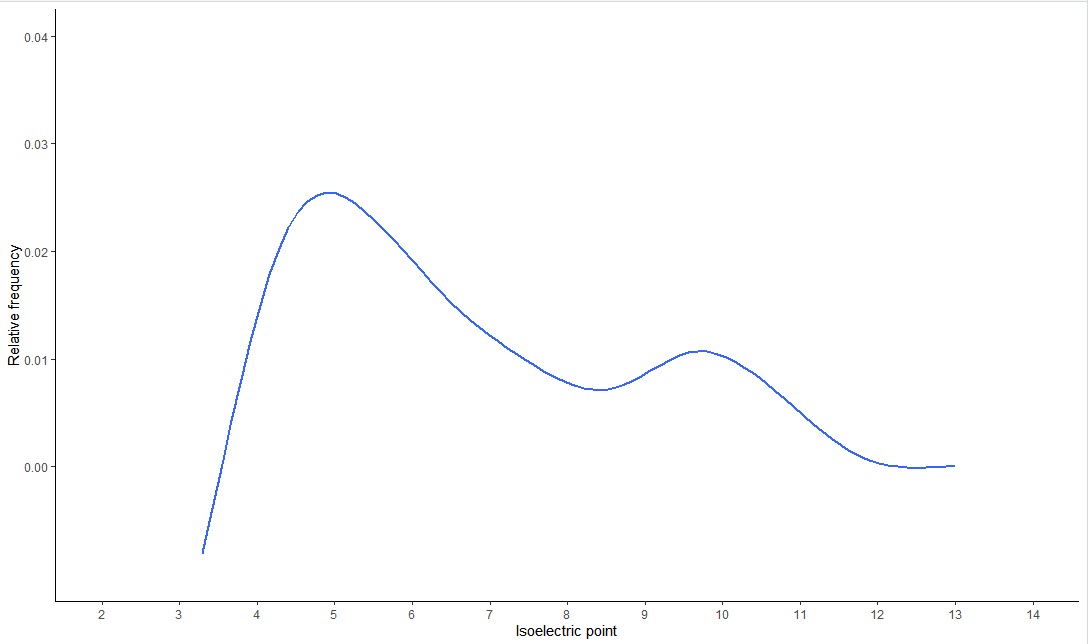
绘制蛋白质组的全局 pIs图
1 | # Step 1 读入数据 |
参考
代码获取
关注公众号“生信之巅”,聊天窗口回复“85d7”获取下载链接。
 |
 |
敬告:使用文中脚本请引用本文网址,请尊重本人的劳动成果,谢谢!